Idt Qpcr Primer Design Tool
The PrimerQuest design tool (www.idtdna.com/SciTools)—one of the most frequently used IDT SciTools® programs—enables customers to design primers and probes for a wide variety of applications. It is the tool of choice for PCR, sequencing, and customized designs. For qPCR assays for human, mouse, or rat targets, we recommend selecting assays using our PrimeTime® qPCR Assay Library Selection Tool; for other species or highly customized designs, use the PrimerQuest tool (see the sidebar, When to use the PrimerQuest Tool vs. the Predesigned qPCR Assay database, below).
Why Use the PrimerQuest Tool?
The advantage of using the PrimerQuest program lies in its customizable designs, which allows users to adjust reaction conditions (e.g., primer, salt, and Mg2+ concentrations), force use of a particular primer or probe sequence, and specify design regions. To view all of the adjustable parameters, simply click Show Custom Design Parameters (Figure 1, red arrow) from the program's main page.
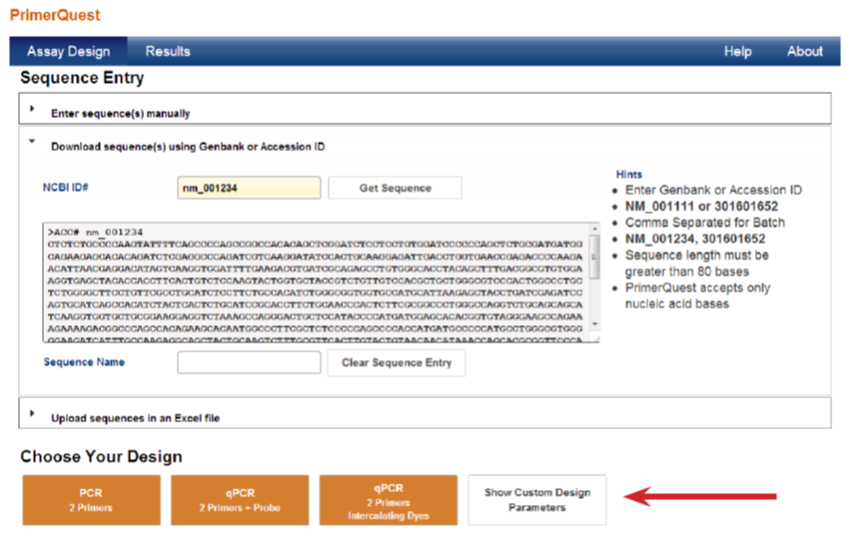
Figure 1. Customize parameters for primer designs.
The parameter page is shown in Figure 2. Click on a parameter name for its definition ("1"). The PrimerQuest tool also has 5 parameter presets ("2") that provide the parameters for each design scenario and create a good starting point for most custom designs. Note that probe parameters are not displayed until qPCR (2 Primers + Probe) ("3") is selected.
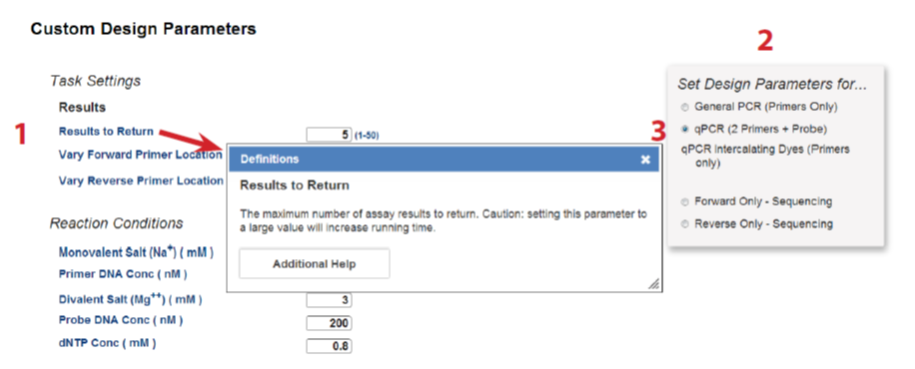
Figure 2. Set reaction parameters for your designs. (1) Hover over the parameter name to get a pop up box with its definition. (2) Choose one of the 5 parameter presets. (3) Select the qPCR (2 Primers + Probe) parameter preset to access the probe parameters.
Switching from intercalating dye assays to probe-based assays
The PrimerQuest tool can help you move from PCR assays that use intercalating dyes to probe-based PCR assays. To identify a probe sequence to go with your primers, simply:
- Enter your target sequence into the Sequence Entry Box.
- Open Custom Design Parameters and select qPCR (2 Primers + Probe) from the parameter presets.
- Scroll down to the Partial Design Input section and enter your forward and reverse primer sequences (Figure 3).
- Click Get Assays.

Figure 3. Identify a probe sequence for your primer set.
Defining primer start/stop locations
The PrimerQuest program can define primer positioning if you require one or both of your primers to start or stop at a specific location. Either the 5' or 3' end of the forward or reverse primer can be easily set by inputting the position within the gene into the Primer Force Location Settings section (Figure 4).
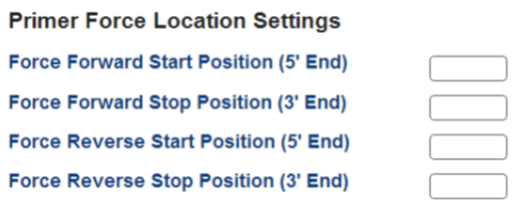
Figure 4. Set specific primer start or stop locations.
Although the tool allows all 4 locations to be set, defining all 4 locations is not necessary. Setting fewer locations provides the tool with more flexibility to produce better designs. Click Get Assays to view designs that meet your location needs.
Overlapping exon junctions
Many PCR applications recommend using assays that cross exon junctions to prevent amplification of genomic DNA. While the PrimerQuest tool cannot pull exon locations from the NCBI database, you can set parameters within the tool to force designs to cross specific locations, such as exon junctions (Figure 5).

Figure 5. Force designs across exon junctions and other specified locations.
Obtain exon locations from the NCBI database. Enter the specified locations into the Overlap Junction List (formatted as a comma separated list), and click Get Assays to see assays that are required to overlap one of these regions. Adjust the minimum overlap of the 5' and 3' ends of the primers to define the amount of flexibility the tool has for that location. IDT recommends increasing the number of Results to Return in order to see assays that cross all specified locations.
Including/excluding design regions
The parameters of the PrimerQuest Tool can also be set to limit primer and probe design to specific regions. Use the Included Region parameter to restrict assay location to within a designated sequence range; e.g., when set to 200−400, primers and probe must fall within base pairs 200−400 (Figure 6). Note that only one included region is allowed per design, and the amplicon size must be smaller than the included region size.
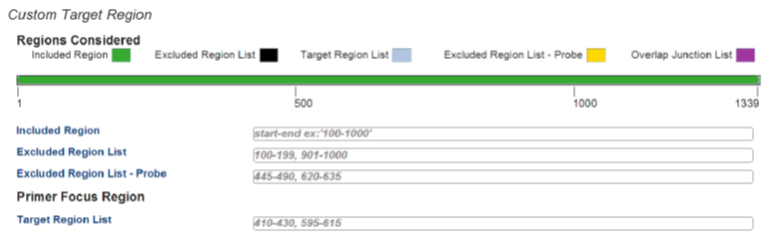
Figure 6. Specify the target region.
The Excluded Region List denotes locations where primers and probe must not bind (Figure 6). For example, if the excluded region is bases 500−1000 for the target selection shown in Figure 6, and the amplicon size is defined as 200 bp in length, the PrimerQuest tool would return designs on either side of the excluded region, within bases 1−500 or 1000−1339. Note that if the specified amplicon size is larger than the excluded region, it may actually span this region, with primers and probe binding on either side of the excluded bases. Multiple excluded regions may be defined per design. This feature can be used to avoid regions of low complexity or high secondary structure, or to more narrowly define where primers cannot reside.
Overall, the PrimerQuest program is the tool of choice for basic PCR designs and customized designs for qPCR and sequencing primers. For any further questions, please contact applicationsupport@idtdna.com.
Idt Qpcr Primer Design Tool
Source: https://www.idtdna.com/pages/education/decoded/article/primerquest-design-tool-from-basic-to-highly-customizable-designs
Posted by: frederickshabligne.blogspot.com

0 Response to "Idt Qpcr Primer Design Tool"
Post a Comment